The ROV is deployed and we’re watching the seabed habitat in real time. Seaweed…a crab…a glimpse of a fish… What if we could find out what animals are actually living in the sea beneath the waves?
We can and we did!
We were on the sail training tall ship Pelican of London for the annual Plymouth Ocean Science Voyage and deployed my ROV* in several locations to compare different seabed habitats. Our trainees were watching the camera footage live on a tablet.
Pelican anchored off Lulworth Cove, Dorset, in 10-15 m depth. The location is open to the English Channel to the south and bounded by chalk cliffs to the north (Figure 1). Apart from ripples in the sandy sediment, the sea bottom was fairly featureless. No seaweeds or rocks, worms or fish. A sole spider crab moved among fragments of crustaceans and empty sea shells. As elsewhere in British shelf seas, we can only suspect bottom trawling responsible for a seabed almost devoid of life.
Within Lulworth Cove, at around 2.5 m depth, a different picture emerged: a dense and highly diverse habitat of kelp, seaweeds, sponges and coralline algae covered a substrate of coarse sand and boulders fallen from the cliffs above. The odd fish flitted across the camera lens. Hydroids cover kelp fronds swaying with the swell – every surface occupied by life.
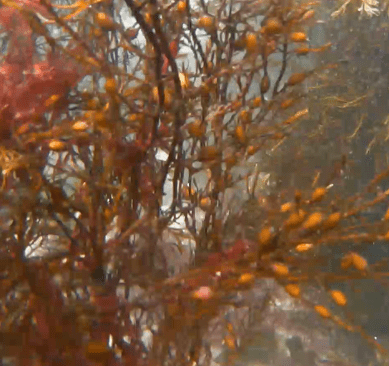
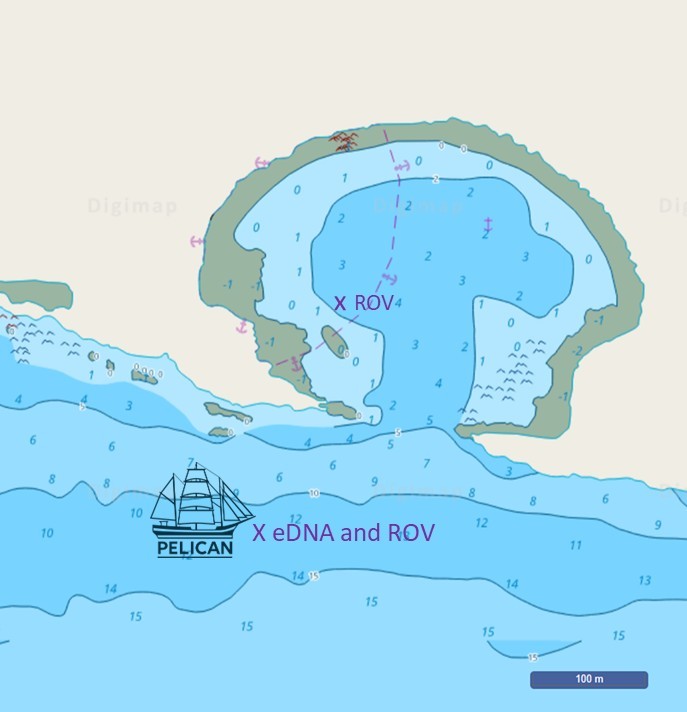
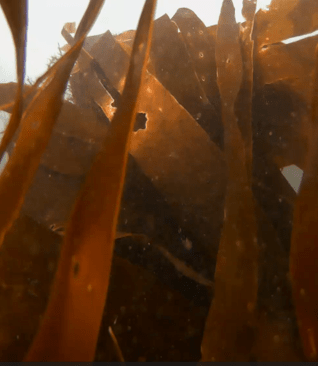


Figure 1: Location where the eDNA sample and underwater footage was taken in and off Lulworth Cove, Dorset. Within Lulworth Cove, the coarse sandy seabed was covered in dense seaweeds of high diversity (top row and bottom right). Outside the cove, in the English Channel, the rippled sandy seabed was almost devoid of life.
To find out what vertebrates live and pass through here, we deployed an inDepth automatic sampling device by Applied Genomics from Pelican on anchor (Figure 1). For 12.25 hours, a full tidal cycle, the device filtered water at 4 m depth for environmental DNA (eDNA) extraction. The results are intriguing:
- Although the seabed was barren at the anchoring site, DNA of a 35 species of fish was detected, 17 of which with a moderate to high certainty. The food web relying on benthic life at this location is damaged, most likely by destructive fishing practices, such as bottom trawling. Nevertheless, nearby coastal shallows, such as Lulworth Cove, will provide nursery grounds for organisms that spill over into more open waters and could re-establish a healthier seabed in no-take zones. The ROV footage also showed organic matter suspended in the water column (marine snow), which indicates nutrient-rich, productive waters.
- Commercially relevant fish detected in the sample included anchovy, sardine, herring, mackerel, whiting, seabass, hake and haddock. In addition, key food-web species, such as conger eel, sand smelt, sandeel and several species of wrasse, indicate a functioning ecosystem. We filtered water over a full tidal cycle, and this means that the sample contains eDNA from waters carried along the coast by the current and tides with velocities of up to 0.6 m/s (Figure 2), covering a range of several hundred meters around the sampling position.
- Three different rays, namely Raja sp., the Spotted ray (Raja montagui) and the Blonde ray (Raja brachyura), were detected at Lulworth. Although these are not yet evaluated by the IUCN, they are of conservation concern in British waters, as they are, as characteristic for elasmobranchii (sharks, skates, rays and sawfish), slow to mature to reproductive age and threatened as bycatch, by unsustainable fishing and by fishing practices that destroy habitat, such as bottom trawling.
- Detected as faint signal only, the presence of the short-snouted seahorse at the anchoring location is unlikely, given the condition of the seabed. Rather, its eDNA was probably transported to the sampling location by the tide from a near-shore environment featuring seagrass or macrophytes (large algae), such as we found inside Lulworth Cove (Figure 1).
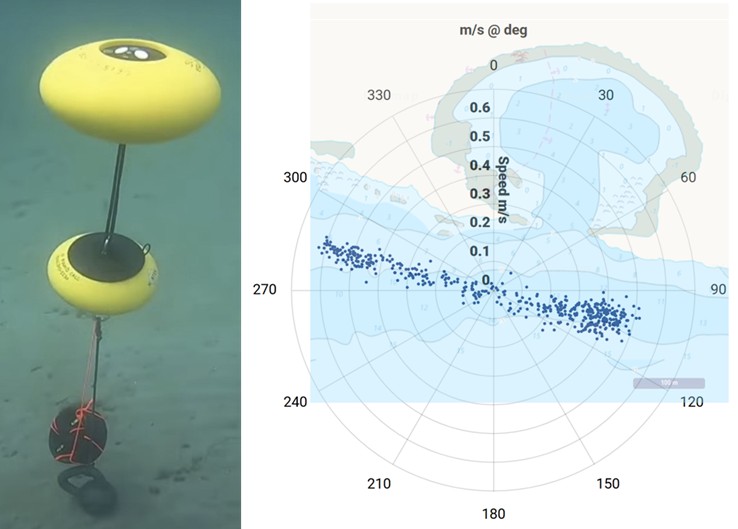
Figure 2: The upward pointing Nortek ECO acoustic Doppler current profiler deployed at the seabed (left, propriety image (c) Nortek). Data collected at 7 m water depth over the full tidal cycle during which eDNA was collected (right). Each blue dot represents a measurement, their distances from the centre indicate velocity measured in m/s and their positions on the compass rose provides their directions. The chart overlay illustrates directional alignment with coastal features. Read more about Nortek ECO in previous blogs (Bob by Nortek Group, The Elegance of Simplicity and The Power of Data).
eDNA analysis takes time and could not be undertaken during the voyage. Therefore, we arranged a webinar in January, during which Sebastian Mynott brought eDNA to life, explaining this state-of-the-art technology not only to participants on the voyage, but also to the biology A-level students at collaborating schools and conservation organisations, such as the Shark Trust.
The full project data, reports and webinar slides are available for download from my Projects page. Feel free to share for educational and scientific purposes, and please respect any copyright statements made on the Applied Genomics report.
*ROV – remotely operated vehicle, model Pioneer, the predecessor of the X1 by blueye Robotics
Acknowledgements
I thank Sea Changers for part-funding the eDNA sampling and analysis, and Nortek Group for their continued support by loaning the current meter free of charge for our educational mission on Pelican of London. Crew and volunteers on Pelican of London deserve a special mention for their support with instrument deployment and science projects on board – integrating science into sail training is a challenge that they have taken on alongside traditional ‘salty’ activities. Finally, many thanks to Sebastian Mynott of Applied Genomics, who brought eDNA to life in a webinar, explaining this state-of-the-art technology, not only to participants on the voyage, but also to the biology A-level students at collaborating schools.

